By
In reinforcement learning (RL), one of the major machine learning (ML) paradigms, an agent interacts with an environment. How well an RL agent can solve a problem, can be sensitive to choices such as the policy network architecture, the training hyperparameters, or the specific dynamics of the environment. A common strategy to deal with this sensitivity, is to first carefully design a neural architecture based on experience and domain-knowledge, followed by an optimization of the training hyperparameters for a specific formulation of the environment. But doing this all manually is not fun and also not really in the spirit of end-to-end optimization. Therefore, for tackling the RNA Design problem (Figure 2), we formulated an automatic reinforcement learning (auto-RL) approach, which searches for the best reinforcement learning formulations by jointly optimizing parameters of the environment (e.g. the shape of the reward function), the neural architecture, and training hyperparameters. The combination of agent (Meta-LEARNA) and environment, which our Auto-RL yielded, achieves new state-of-the-art results, while also being up to 1765 times faster in reaching the previous state-of-the-art performance for the problem of RNA design (Figure 1).
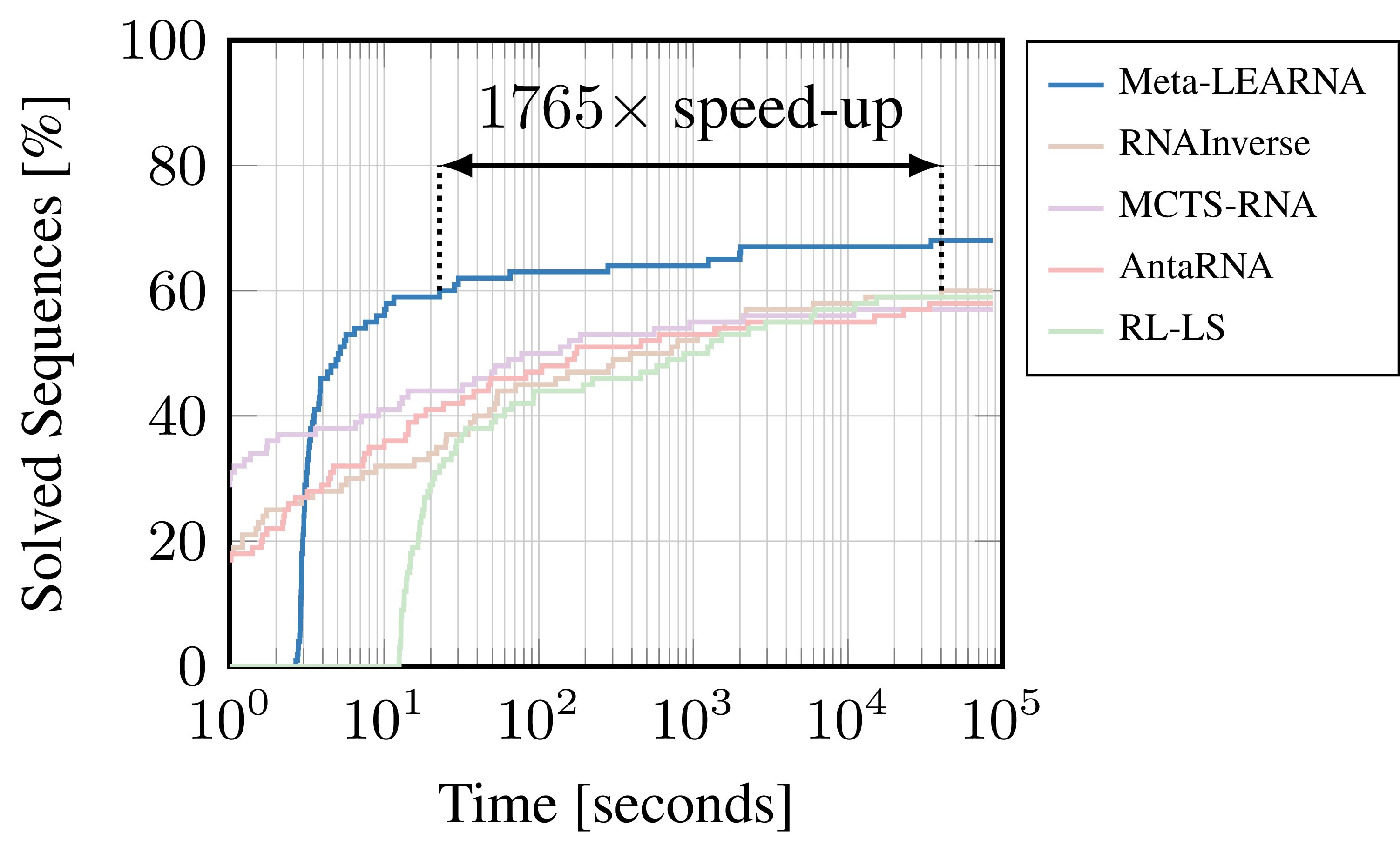
Figure 1: Comparison of our best reinforcement learning formulation (Meta-LEARNA) and state-of-the-art algorithms (RNAInverse, MCTS-RNA, antaRNA, RL-LS) on one of the most commonly used RNA Design benchmarks (Eterna100).
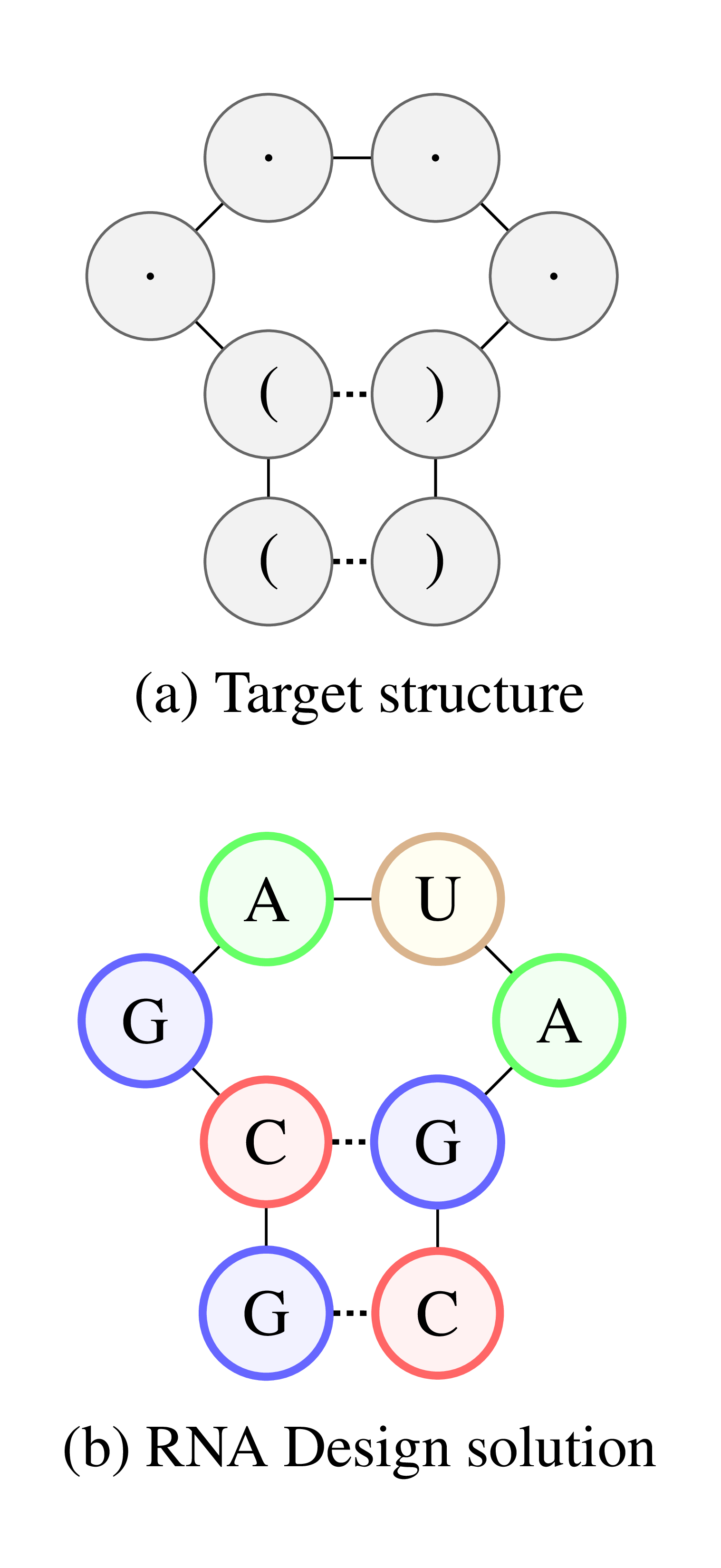
Figure 2: lllustration of the RNA Design problem. (a) Target structure in dot-bracket notation. (b) RNA sequence that satisfies the structural constraints.
Why design RNA Molecules?
Along with DNA and proteins, RNA is one of the three major macromolecules, which are essential for all forms of life. Although mostly known for its role during protein synthesis, RNA can regulate biological processes directly and has been connected to diseases such as autism, Parkinson’s, Alzheimer’s and epilepsy. As with proteins, the regulatory power of RNA can be exploited to achieve certain functions in the body. The functional properties of an RNA depend highly on its folding. Therefore, to engineer an RNA molecule with a desired function, a possible approach is to first construct a corresponding folding (Figure 2a) and then design a sequence which satisfies this folding (Figure 2b).
Reinforcement Learning for Designing RNA Molecules
Designing RNA molecules which satisfy certain constraints is a hard and time-consuming process, therefore research into automated approaches thrived since the first algorithm proposed by et al. Even though people do not usually use ML for this problem, we decided to develop a novel generative deep reinforcement learning approach for RNA Design. Our RL agent reads one structural element at a time, and chooses the corresponding nucleotides. Once all sites are assigned nucleotides, the resulting RNA sequence is folded, and the agent receives a reward based on how well the folded sequence and the target structure match (Figure 3). To not start from scratch for each new target folding, we meta-learn across many RNA sequences. This allows our system to learn the dynamics underlying RNA Design and to perform similarly well across all considered sequence lengths.
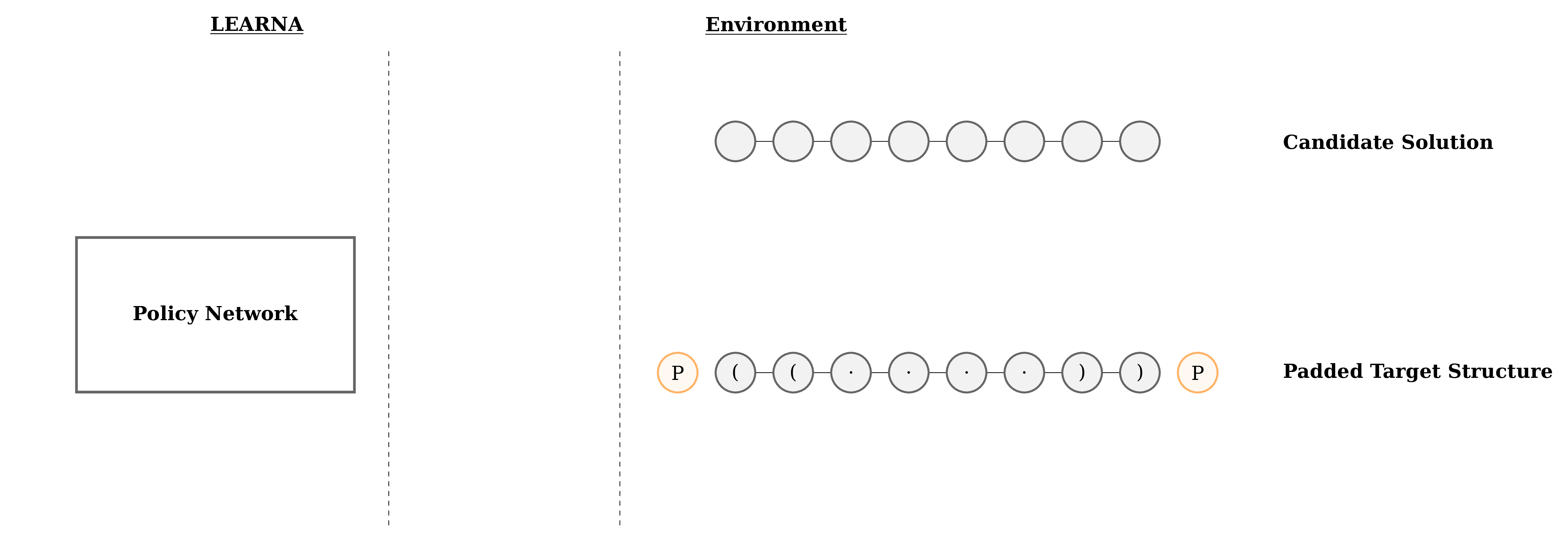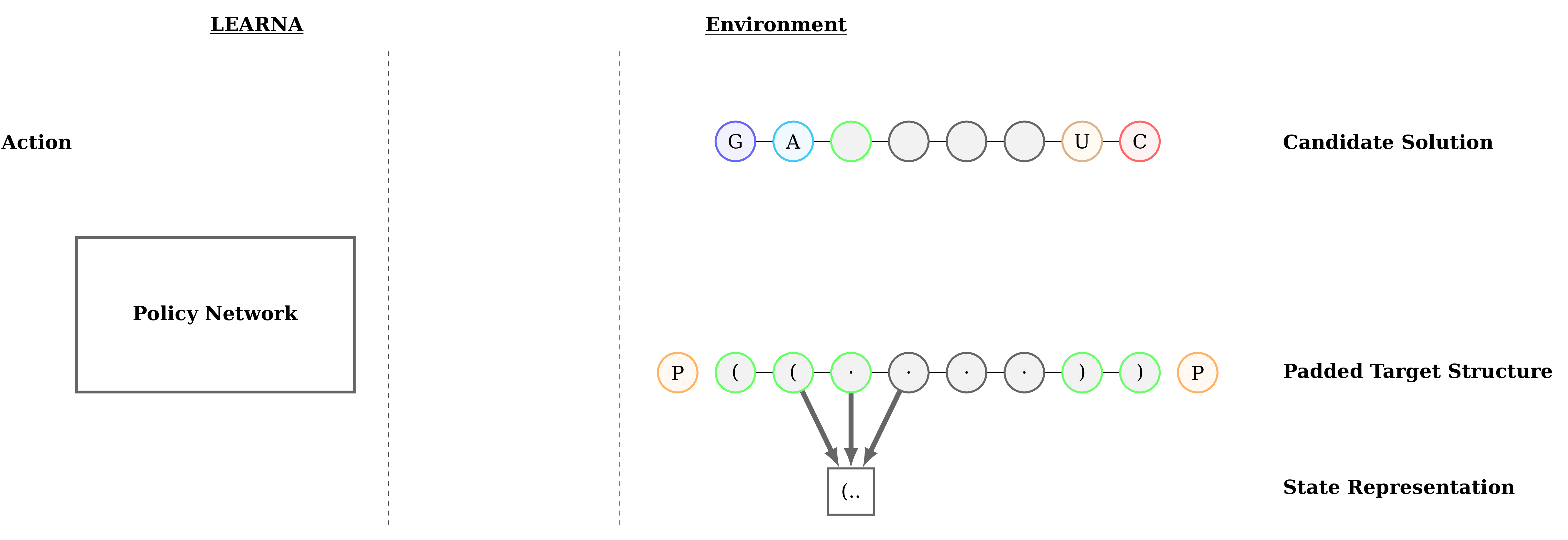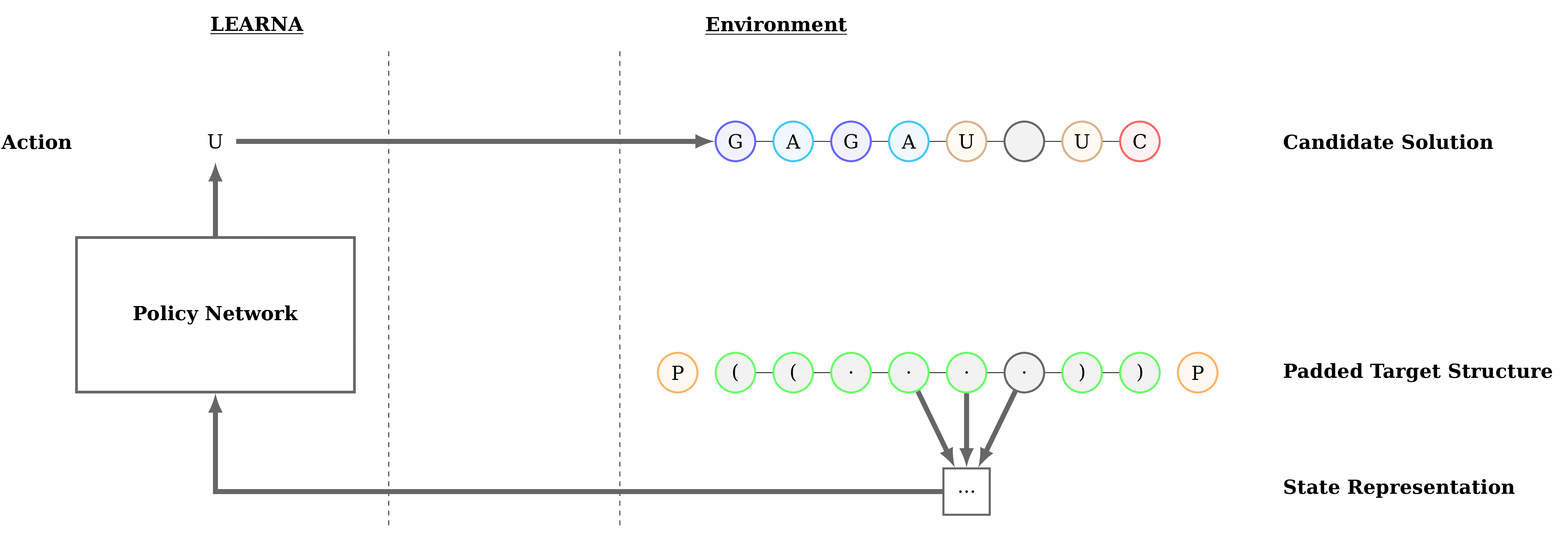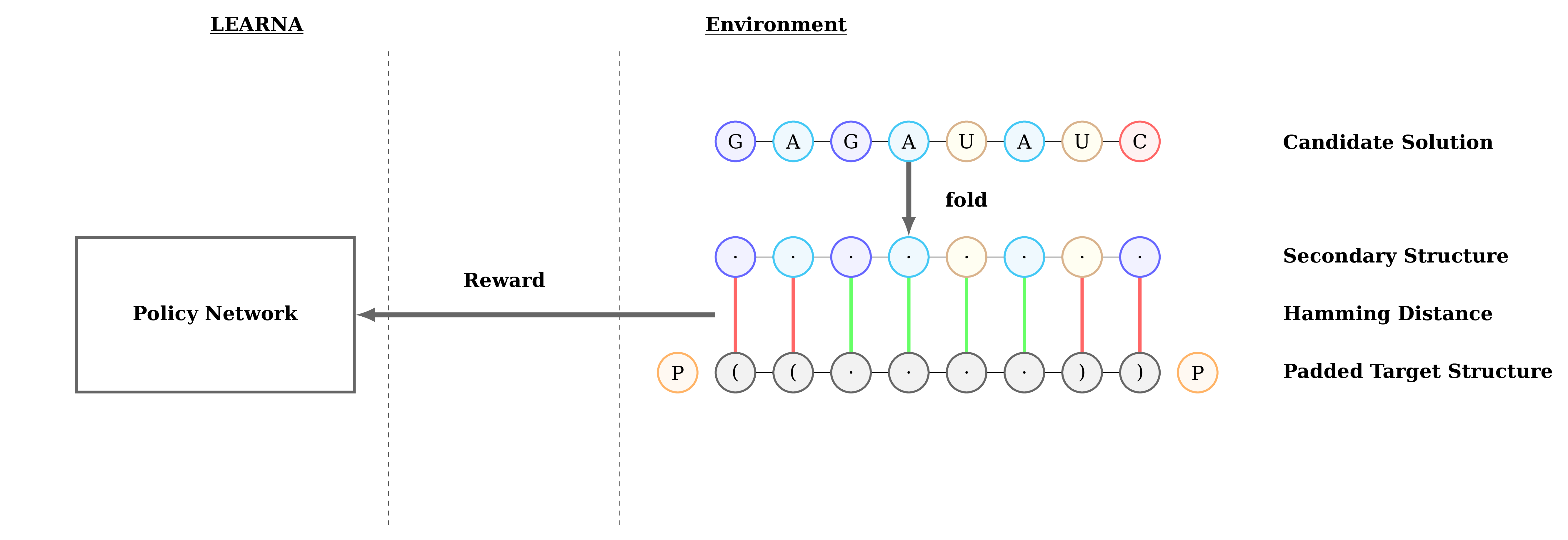
Figure 3: Interactions between the agent (LEARNA) and the environment. The padded target structure serves as a template for the states which encode the sequence information via a n-gram centered around the current site. Once LEARNA is provided with a state, it chooses an action to place a nucleotide or a pair of nucleotides. Finally, when all sites are assigned nucleotides, the candidate solution is folded and the environment produces the reward based on the Hamming Distance between the folded candidate solution and the target structure.
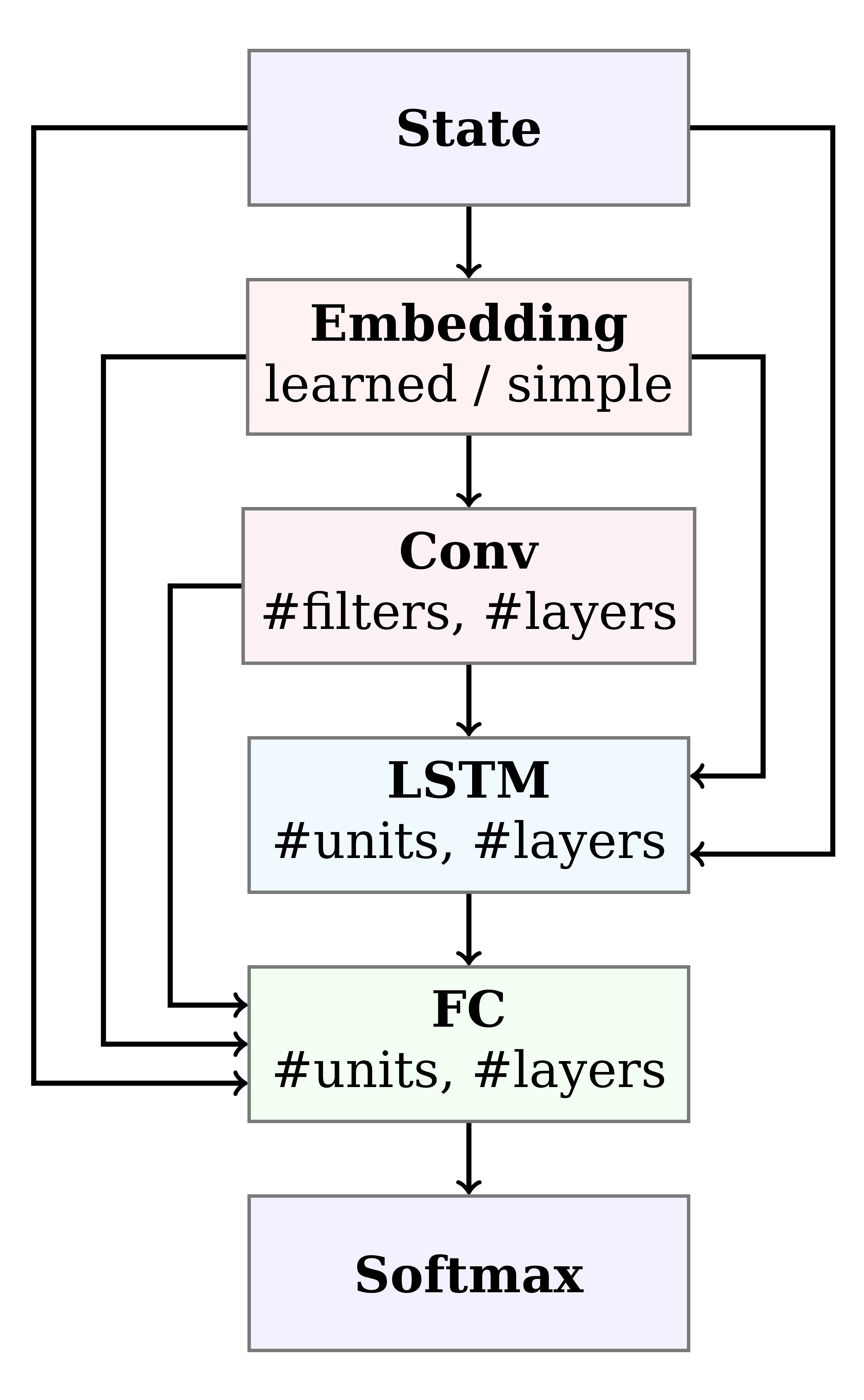
Figure 4: Architecture search space. Paths correspond to architectures. Note that our search space includes elements of recurrent neural networks (RNNs) and convolutional neural networks (CNNs).
Auto-RL for applying RL to a novel problem setting
Since RNA Design is a novel problem setting for reinforcement learning, it was not clear in advance how to best formulate the training pipeline, environment, and neural architecture. To optimize over possible formulations, we automatically and jointly searched over these domains, using the recently published optimizer BOHB. In our 14-dimensional search space, we considered different reward shapes, state formulations, training hyperparameters, and a multitude of different neural architectures including recurrent and convolutional building blocks (Figure 4). Further, we used fANOVA to show that some choices in all three categories were important; this result highlights how crucial this joint search was to the success of our approach.
Conclusion
We proposed a generative deep reinforcement learning algorithm for the RNA Design problem, which constructs candidate solutions in an end-to-end fashion. By automatically optimizing all parts of our reinforcement learning formulation jointly, we achieve new state of the art results on the two most commonly used RNA Design benchmarks, while also being much faster than all previous algorithms for this problem. Since our novel automatic reinforcement learning (auto-RL) approach worked well for this task, we hope to see auto-RL be further explored and applied to other problems. Our source code and data is freely available and we refer the interested reader to our ICLR 2019 publication or our podcast interview at This Week in Machine Learning & AI (TWiML & AI) about automated ML for RNA Design with Danny Stoll. Also see our other recent NAS papers.